Tutorial
Table of Contents #
How to Setup and Compile #
Note that this tutorial is based on latest version (Helios Service Release 1, Build id: 20100917-0705) of Eclipse IDE.
If you are using older version, you may see somewhat different screen. Please update your Eclipse in that case.
As mentioned above, you can apply more than one option such like
The table below lists algorithm code and parameters.
* Similarity method should be one of {pearson, cosine, msd, mad, invuserfreq}.
** Similarity prefetch file should be used together with corresponding train/test split file for accurate test.
*** log_mult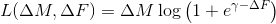 ,
log_add
,
log_add 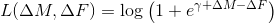 ,
,
exp_mult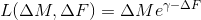 ,
exp_add
,
exp_add 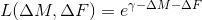 ,
,
hinge_mult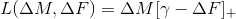 ,
hinge_add
,
hinge_add  ,
,
abs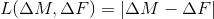 ,
sqr
,
sqr 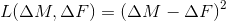 ,
,
expreg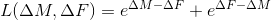 ,
,
l1reg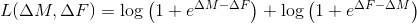
Setting up with Eclipse
- Download Eclipse Project file.
- Run Eclipse, and select "File > Import". You will see the following window.
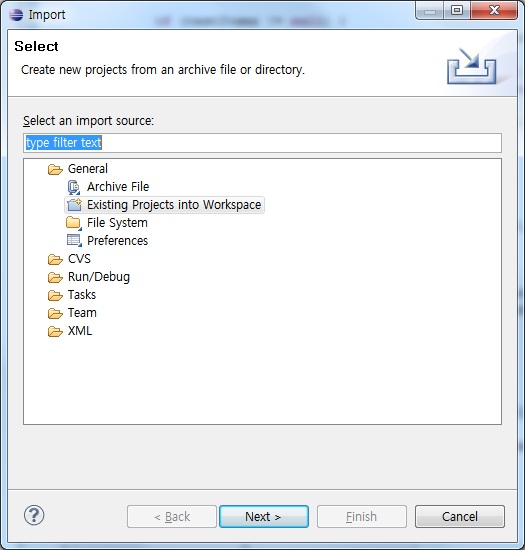
- Choose "Existing Projects into Workspace", then press "Next" button. You will see the following window.

- Choose a radio button "Select archive file:", and select the zip file downloaded by step 1.
- Make sure that both "Prea" and "UJMP_src" in the "Projects:" box are checked.
- Press the button "Finish". Then, you can see that the project is added in your workspace.
- (If you do not use auto-compile option) Select "Project > Build Project" to compile the project.
- Download dataset from Download section and unzip them in the Prea directory.
Compiling on Command Line
- Download source code file.
- Unzip the downloaded file in a directory, and locate yourself in the same directory.
- Run the next command. This automatically compiles all files needed to run this program.
> javac prea/main/Prea.java* Note that we have two files containing main method. RankBasedSVD and PairedGlobalLLORMA are executed with PreaRank, and all other methods are done with Prea.
> javac prea/main/PreaRank.java
How to Run #
Running the Program
Make sure that you successfully compiled the source code, or have executables in the directory.
First, you can run the program without specifying anything. In this case, the program runs all algorithms with a default demo data file "netflix_3m1k", bundled with the install file.
* This number contains the number of data points used for validation set.
First, you can run the program without specifying anything. In this case, the program runs all algorithms with a default demo data file "netflix_3m1k", bundled with the install file.
> java prea/main/Prea
For more advanced use, you can specify three kinds of arguments.
More than one argument can be used simultaneously, independent to each other.
For an argument which was not specified, the default value in the table below would be applied.
Detailed explanation about data split can be found in Documentation section.
| Options | Syntax | Default value | Example |
| Input file | -f [File Name] | netflix_3m1k | java Prea -f movieLens_1M |
| Data Split | -s simple [Test ratio] | simple, 0.2 | java Prea -s simple 0.2 |
| -s pred [Split file name] | java Prea -s pred netflix_3m1k_split.txt | ||
| -s kcv [Fold count] | java Prea -s kcv 5 | ||
| -s rank [Train point per user]* | java Prea -s rank 20 | ||
| Algorithm | -a [Algorithm code. See below.] | (Run all.) | java Prea -a regsvd [Parameters of the algorithm. See below.] |
> java prea/main/Prea -s kcv 5 -f movieLens_100K
- [data file]: the name of data file, without the file extension.
Ex) netflix_3m1k for netflix_3m1k.arff.
> java prea/main/PreaRank -s rank 20 -f movieLens_100K
The table below lists algorithm code and parameters.
| Algorithm | Keyword | Parameter List Example |
| Constant | const | java prea/main/Prea -a const |
| Overall Average | allavg | java prea/main/Prea -a allavg |
| User Average | useravg | java prea/main/Prea -a useravg |
| Item Average | itemavg | java prea/main/Prea -a itemavg |
| Random | random | java prea/main/Prea -a random |
| User-based CF | userbased | java prea/main/Prea -a userbased [neighbor size(k)] [similarity method]* [(optional) default] [default value] [(optional) usersim]** [user similarity prefetch file name] |
| java prea/main/Prea -a userbased 50 pearson java prea/main/Prea -a userbased 50 pearson default 3.0 java prea/main/Prea -s pred netflix_3m1k_split.txt -a userbased 50 mad default 3.0 usersim netflix_3m1k_userSim.txt | ||
| Item-based CF | itembased | java prea/main/Prea -a itembased [neighbor size(k)] [similarity method]* [(optional) default] [default value] [(optional) itemsim]** [item similarity prefetch file name] |
| java prea/main/Prea -a itembased 50 cosine java prea/main/Prea -a itembased 50 cosine default 3.0 java prea/main/Prea -s pred netflix_3m1k_split.txt -a itembased 50 msd default 3.0 itemsim movielens_1M_itemSim.txt | ||
| Slope One | slopeone | java prea/main/Prea -a slopeone |
| Regularized SVD | regsvd | java prea/main/Prea -a regsvd [feature count] [learning rate] [regularizer] [max iteration] |
| java prea/main/Prea -a regsvd 10 0.005 0.1 200 | ||
| NMF | nmf | java prea/main/Prea -a nmf [feature count] [regularizer] [max iteration] |
| java prea/main/Prea -a nmf 100 0.0001 5 | ||
| PMF | pmf | java prea/main/Prea -a pmf [feature count] [learning rate] [regularizer] [momentum] [max iteration] |
| java prea/main/Prea -a pmf 10 50 0.4 0.8 20 | ||
| Bayesian PMF | bpmf | java prea/main/Prea -a bpmf [feature count] [max iteration] |
| java prea/main/Prea -a bpmf 2 20 | ||
| Nonlinear PMF | nlpmf | java prea/main/Prea -a nlpmf [feature count] [learning rate] [momentum] [max iteration] [kernel inverse width] [kernel variance RBF] [kernel variance bias] [kernel variance white] |
| java prea/main/Prea -a nlpmf 10 0.0001 0.9 2 1 1 0.11 5 | ||
| Fast NPCA | npca | java prea/main/Prea -a npca [validation ratio] [max iteration] |
| java prea/main/Prea -a npca 0.15 5 | ||
| Rank-based CF | rank | java prea/main/Prea -a rank [kernel width] |
| java prea/main/Prea -a rank 1.0 | ||
| Singleton Global LLORMA | sgllorma | java prea/main/Prea -a sgllorma [feature count] [learning rate] [regularizer] [max iteration] [model count] |
| java prea/main/Prea -a sgllorma 5 0.1 0.001 100 50 | ||
| Singleton Parallel LLORMA | spllorma | java prea/main/Prea -a spllorma [feature count] [learning rate] [regularizer] [max iteration] [model count] [thread count] |
| java prea/main/Prea -a spllorma 10 0.01 0.001 100 50 8 | ||
| Rank-based SVD | ranksvd | java prea/main/Prea -a ranksvd [loss code]*** [local model rank] [learning rate] |
| java prea/main/Prea -a ranksvd log_mult 1 1500 | ||
| Paired Global LLORMA | pgllorma | java prea/main/Prea -a pgllorma [loss code]*** [local model count] [local model rank] [learning rate] |
| java prea/main/Prea -a pgllorma log_mult 5 1 1500 |
** Similarity prefetch file should be used together with corresponding train/test split file for accurate test.
*** log_mult
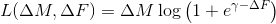 ,
log_add
,
log_add 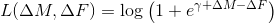 ,
,exp_mult
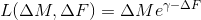 ,
exp_add
,
exp_add 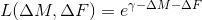 ,
,hinge_mult
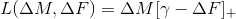 ,
hinge_add
,
hinge_add  ,
,abs
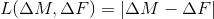 ,
sqr
,
sqr 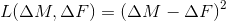 ,
,expreg
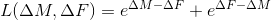 ,
,l1reg
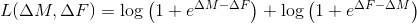
How to Read Result
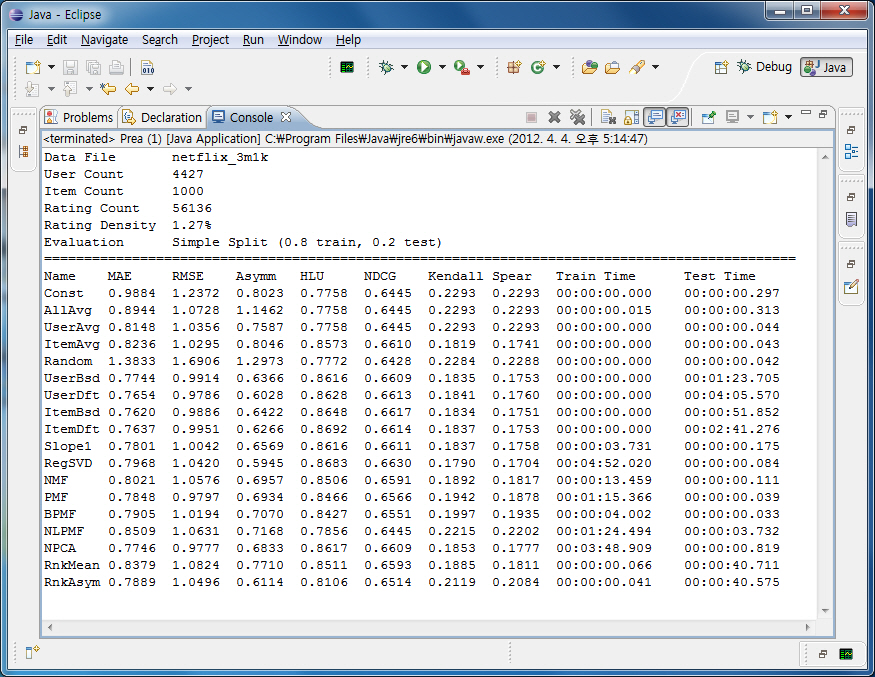
The experimental result is shown in console with plain text.
First, it shows properties of the data file, including file name, user/item count, and rating density.
Each row represents each algorithm, and each column lists various evaluation metrics.
Note that for HLU and NDCG, higher values mean better performance, while lower values are better for all other metrics.
The following figure shows an example of execution with Netflix Tiny set.

Tip: Increasing Heap Memory
As the data file grows bigger, this program requires lots of memory space. You may encounter the following message:
Exception in thread "main" java.lang.OutOfMemoryError: Java heap space
In this case, the following Java option can be useful:
> java -Xms1024m -Xmx2048m Prea ...
This option means heap size is initially set to 1024MB (1GB), and can be expanded upto 2048MB (2GB).
You may set both size equally.
Using in Other Tools #
PREA provides wrapper functions for other popular tools such as Matlab.
With these, it is possible to conduct an experiment by calling PREA in other tools.
Using in Matlab
- Download source code file, and unzip it in a directory.
- Locate the Matlab current directory to the directory. In GUI version, you can use the following red box.
Make sure that there are all executable files and two matlab files (matlab2arff.m, run.m).
- Load the data file in Matlab. The matrix should in the format of users (row) by items (column). If it is reversed, it is necessary to transpose it at this point.
- Run the wrapper function with the following formats.
- The simplest format specifies only matrix name in Matlab. In this case, it runs constant model (Median) as a default. For example,
prea(movielens);
- You can specify the output ARFF file from the Matlab matrix, if you want to use it again later.
For example, the following code will create "movielens.arff" file in the directory.
prea(movielens, 'movielens');
- With another parameter, you can specify a CF algorithm keyword to be used. For example,
prea(movielens, 'movielens', 'itembased');
prea(movielens, 'movielens', 'bpmf');
prea(movielens, 'movielens', 'npca');
- Lastly, you can specify arguments for each algorithm. This takes two steps; first, you should make "options" with parameters you want to specify,
then, add it as a parameter to "run" function. (Without specifying these parameters just as the previous case, default parameters are used.)
options = struct('feature_count', 5, 'maxiter', 20);The list of available options for each algorithm is as follows:
prea(movielens, 'movielens', 'bpmf');
Algorithm Keyword Parameter List Constant const None Overall Average allavg None User Average useravg None Item Average itemavg None Random random None User-based CF userbased neighbor_size Item-based CF itembased neighbor_size Slope One slopeone None Regularized SVD regsvd feature_count, learning_rate, regularizer, maxiter NMF nmf feature_count, learning_rate, maxiter PMF pmf feature_count, learning_rate, regularizer, momentum, maxiter Bayesian PMF bpmf feature_count, maxiter Nonlinear PMF nlpmf feature_count, learning_rate, momentum, maxiter, kernel_inverse_width, kernel_var_rbf, kernel_var_bias, kernel_var_white Fast NPCA npca validation_ratio, maxiter Rank-based CF rank kernel_width Singleton Global LLORMA sgllorma feature_count, learning_rate, regularizer, maxiter, maxmodel Singleton Parallel LLORMA spllorma feature_count, learning_rate, regularizer, maxiter, maxmodel, thread
- The simplest format specifies only matrix name in Matlab. In this case, it runs constant model (Median) as a default. For example,
- The output from the toolkit is shown in Matlab screen.